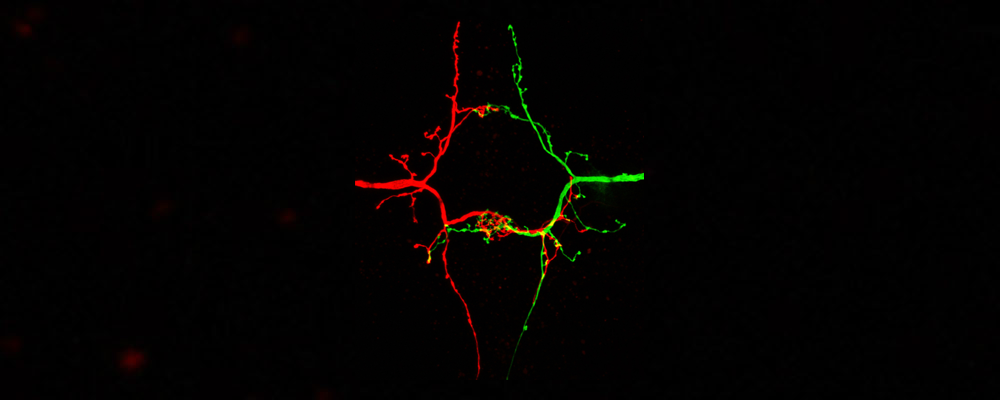
Neuronal Wiring
1) Molecular Diversity and Function of Membrane Receptors:
In a central project in the lab we have been using the model organism Drosophila investigating the recognition specificity and signal transduction of membrane receptors during developmental wiring of the nervous system. Specifically, we have been studying the function of the Ig-domain containing neuronal receptor “Dscam”. The Drosophila Dscam1 receptor is closely related to the human protein Down syndrome cell adhesion molecule (DSCAM). Through alternative splicing the Drosophila Dscam1 gene (but not vertebrate DSCAM) gives rise to thousands of diverse cell surface receptors (~38,000) thought to provide homophilic and possibly heterophilic recognition specificity for neuronal wiring and immune responses (Schmucker et al. 2000, Cell; Watson et al. Science, 2005; Chen et al. Cell; 2006; Meijers et al. Nature 2007).
In this context we have been investigating the in vivo role of Dscam1’s isoform specificity during neuronal wiring. Genetic studies reveal that reduction of Dscam1 diversity strongly impairs neurite branching. Furthermore, cell-intrinsic functions of Dscam in neurons control complex axon and dendrite morphogenesis (Chen et al. Cell; 2006; Hughes et al. Neuron 2007; He et al. Science 2014; Dascenco et al. Cell, 2016). In this context, isoform-specific homophilic interactions of receptors positioned on interacting membrane compartments result in neurite repulsion. Isoform specificity of Dscam1 provides a form of molecular identity recognition that is used to distinguish self and non-self. Overall, we are interested to dissect the molecular mechanisms of such receptor-based identity recognition and investigate whether this novel concept is of general importance for neuronal wiring.
2) Molecular Organization, expression and function of vertebrate DSCAM and clustered protocadherins (cPcdh) in Xenopus tropicalis
Under construction…
3) Novel molecular and developmental mechanisms of axonal branching
Under construction…
4) Specificity of CNS synaptogenesis:
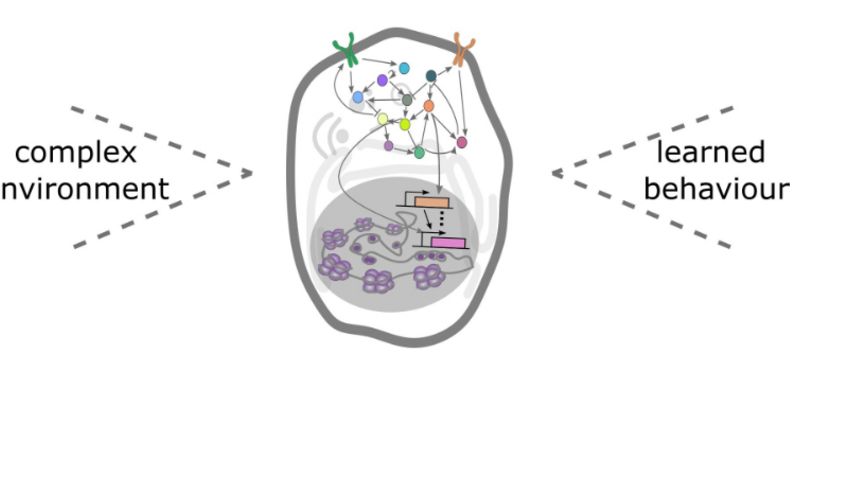
Precise synaptic wiring of the highly complex central nervous system (CNS) underlies cognition, learning, memory, and complex behavior of higher animals. Genetic association studies in humans have linked neurological syndromes (including autism, schizophrenia etc) to mutations in genes potentially controlling CNS wiring, in particular with synapse formation and function (Gilman et al. 2011). But it is extremely difficult to establish any causative links between candidate genes and potential synaptic defects due to the high technical challenges. Characterizing synapse specificity in vivo not only requires imaging at high resolution (including EM studies), but also identification of defined synapses and comparing them across individuals. Especially the latter is difficult to achieve in most experimental systems. In general, it is difficult to diagnose morphological alterations of specific synapses, although such defects, even when subtle, may have profound effects on circuit functions. Indeed, mutations in synaptic components that are linked to human syndromes often do not disrupt gross number or distribution of synapses, but affect only specific morphological characteristics or a subset of synapses (Suedhof, 2008).
It would thus be highly desirable to have a labeling system for single defined central synapses, which allows imaging at the ultrastructural level and simultaneously addressing genetically the contribution of individual molecules in synapse formation and specificity.
In order to identify novel components of the genetic and molecular network underlying CNS synapse formation, we are performing genetic screens in model systems like Drosophila and subsequently characterize function of candidate genes (Urwyler et al. Development, 2018) in different systems. For example, several pro- and anti-synaptogenic factors were identified in the lab in a reverse genetic screen targeting the majority of Drosophila kinases and phosphatases. One of the pro-synaptogenic factors identified is Drosophila Prl-1 (Phosphatase of regenerating liver), which could be shown to act as a membrane anchored, axon-intrinsic factor required for axon arborization and synapse stabilization in a dedicated neuronal compartment (Urwyler et al. Science, 2019). Importantly, the Prl-1 phosphatase functions upstream of AKT in the Insulin signaling pathway where it controls branching and synaptogenesis in a spatially restricted fashion. Remarkably, local restriction of Prl-1 activity in the contralateral axon collateral seems to rely on mRNA-directed control of Prl-1 transport or local translation in this compartment. Ongoing projects address the mechanistic basis for this regulation and functional consequences with respect to neuronal wiring in Drosophila and vertebrates. As an important tool to visualize axonal transport and subcellular RNA localization, we use high resolution confocal im-aging of fluorescently tagged RNA molecules in vivo.